Bootstrap simulation for model prediction
Ewen Harrison
Source:vignettes/bootstrap.Rmd
bootstrap.RmdI’ve always been a fan of converting model outputs to real-life quantities of interest. For example, I like to supplement a logistic regression model table with predicted probabilities for a given set of explanatory variable levels. This can be more intuitive than odds ratios, particularly for a lay audience.
For example, say I have run a logistic regression model for predicted 5 year survival after colon cancer. What is the actual probability of death for a patient under 40 with a small cancer that has not perforated? How does that probability differ for a patient over 40?
I’ve tried this various ways. I used Zelig for a while including here, but it started trying to do too much and was always broken (I updated it the other day in the hope that things were better, but was met with a string of errors again).
I also used rms, including here (checkout the nice plots!). I like it and respect the package. But I don’t use it as standard and so need to convert all the models first, e.g. to lrm. Again, for my needs it tries to do too much and I find datadist awkward.
Thirdly, I love Stan for this, e.g. used in this paper. The generated quantities block allows great flexibility to simulate whatever you wish from the posterior. I’m a Bayesian at heart will always come back to this. But for some applications it’s a bit much, and takes some time to get running as I want.
I often simply want to predict y-hat from lm and glm with
bootstrapped intervals and ideally a comparison of explanatory levels
sets. Just like sim does in Zelig. But I want
it in a format I can immediately use in a publication.
Well now I can with finalfit
There’s two main functions with some new internals to help expand to other models in the future.
Make sure you are on the most up-to-date version of finalfit.
install.packages("finalfit")Create new dataframe of explanatory variable levels
ff_newdata (alias: finalfit_newdata) is
used to generate a new dataframe. See the next section for an
alternative approach to doing this. I usually want to set 4 or 5
combinations of x levels and often find it difficult to get
this formatted correctly to use with predict. Pass the
original dataset, the names of explanatory variables used in the model,
and a list of levels for these. For the latter, they can be included as
rows or columns. If the data type is incorrect or you try to pass factor
levels that don’t exist, it will fail with a useful warning.
library(finalfit)
explanatory = c("age.factor", "extent.factor", "perfor.factor")
dependent = 'mort_5yr'
colon_s %>%
finalfit_newdata(explanatory = explanatory, newdata = list(
c("<40 years", "Submucosa", "No"),
c("<40 years", "Submucosa", "Yes"),
c("<40 years", "Adjacent structures", "No"),
c("<40 years", "Adjacent structures", "Yes") )) -> newdata
newdata
#> age.factor extent.factor perfor.factor
#> 1 <40 years Submucosa No
#> 2 <40 years Submucosa Yes
#> 3 <40 years Adjacent structures No
#> 4 <40 years Adjacent structures Yes
ff_expand() for creating new data frame
ff_expand() has been added to aid in setting up new data
frames for model prediction. It uses two further functions
(ff_mode() and summary_df()) to generate a
data frame which includes the mode of factors (most frequent occurrence
of a factor level) and the mean or median of numeric variables, together
with an expansion of specified factors to include all combinations of
their levels.
This is commonly needed when simulating model predictions.
library(dplyr)
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
colon_s %>%
select(-hospital) %>%
ff_expand(age.factor, sex.factor)
#> # A tibble: 6 × 31
#> age.factor sex.factor id rx sex age obstruct perfor adhere nodes
#> <fct> <fct> <dbl> <fct> <dbl> <dbl> <dbl> <dbl> <dbl> <dbl>
#> 1 <40 years Female 465 Obs 0.521 59.8 0.194 0.0291 0.145 3.66
#> 2 <40 years Male 465 Obs 0.521 59.8 0.194 0.0291 0.145 3.66
#> 3 40-59 years Female 465 Obs 0.521 59.8 0.194 0.0291 0.145 3.66
#> 4 40-59 years Male 465 Obs 0.521 59.8 0.194 0.0291 0.145 3.66
#> 5 60+ years Female 465 Obs 0.521 59.8 0.194 0.0291 0.145 3.66
#> 6 60+ years Male 465 Obs 0.521 59.8 0.194 0.0291 0.145 3.66
#> # ℹ 21 more variables: status <dbl>, differ <dbl>, extent <dbl>, surg <dbl>,
#> # node4 <dbl>, time <dbl>, rx.factor <fct>, obstruct.factor <fct>,
#> # perfor.factor <fct>, adhere.factor <fct>, differ.factor <fct>,
#> # extent.factor <fct>, surg.factor <fct>, node4.factor <fct>,
#> # status.factor <fct>, loccomp <dbl>, loccomp.factor <fct>, time.years <dbl>,
#> # mort_5yr <fct>, age.10 <dbl>, mort_5yr.num <dbl>Run bootstrap simulations of model predictions
boot_predict takes standard lm and
glm model objects, together with finalfit
lmlist and glmlist objects from fitters,
e.g. lmmulti and glmmulti. In addition, it
requires a newdata object generated from
ff_newdata. If you’re new to this, don’t be put off by all
those model acronyms, it is straightforward.
colon_s %>%
glmmulti(dependent, explanatory) %>%
boot_predict(newdata,
estimate_name = "Predicted probability of death",
R=100, boot_compare = FALSE,
digits = c(2,3))
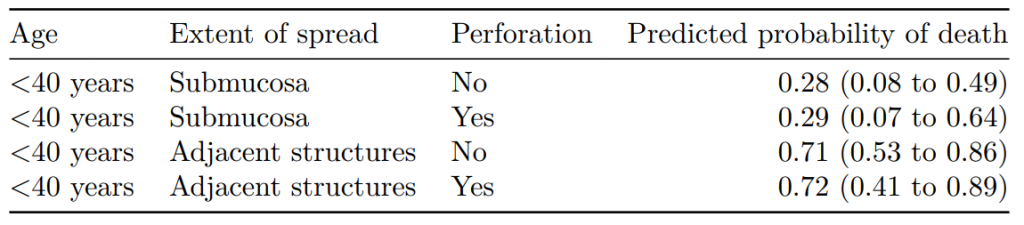
#> Age Extent of spread Perforation Predicted probability of death
#> 1 <40 years Submucosa No 0.28 (0.06 to 0.55)
#> 2 <40 years Submucosa Yes 0.29 (0.05 to 0.56)
#> 3 <40 years Adjacent structures No 0.71 (0.55 to 0.89)
#> 4 <40 years Adjacent structures Yes 0.72 (0.40 to 0.87)Note that the number of simulations (R) here is low for
demonstration purposes. You should expect to use 1000 to 10000 to ensure
you have stable estimates.
Output to Word, PDF, and html via RMarkdown
Simulations are produced using bootstrapping and everything is tidily
outputted in a table/dataframe, which can be passed to
knitr::kable.
Place this chunk in an .Rmd file:
Make comparisons
Better still, by including boot_compare=TRUE (default),
comparisons are made between the first row of newdata and each
subsequent row. These can be first differences (e.g. absolute risk
differences) or ratios (e.g. relative risk ratios). The comparisons are
done on the individual bootstrap predictions and the distribution
summarised as a mean with percentile confidence intervals (95% CI as
default, e.g. 2.5 and 97.5 percentiles). A p-value is generated on the
proportion of values on the other side of the null from the mean,
e.g. for a ratio greater than 1.0, p is the number of bootstrapped
predictions under 1.0. Multiplied by two so it is two-sided.
colon_s %>%
glmmulti(dependent, explanatory) %>%
boot_predict(newdata,
estimate_name = "Predicted probability of death",
#compare_name = "Absolute risk difference",
R=100, digits = c(2,3))
#> Age Extent of spread Perforation Predicted probability of death
#> 1 <40 years Submucosa No 0.28 (0.09 to 0.54)
#> 2 <40 years Submucosa Yes 0.29 (0.05 to 0.63)
#> 3 <40 years Adjacent structures No 0.71 (0.52 to 0.91)
#> 4 <40 years Adjacent structures Yes 0.72 (0.46 to 0.93)
#> Difference
#> 1 -
#> 2 -0.00 (-0.11 to 0.17, p=0.960)
#> 3 0.43 (0.19 to 0.64, p<0.001)
#> 4 0.41 (0.14 to 0.67, p<0.001)What is not included?
It doesn’t yet include our other common models, such as
coxph which I may add in. It doesn’t do lmer
or glmer either. bootMer works well
mixed-effects models which take a bit more care and thought, e.g. how
are random effects to be handled in the simulations. So I don’t have
immediate plans to add that in, better to do directly.
Plotting
Finally, as with all finalfit functions, results can be produced as
individual variables using condense == FALSE. This is
particularly useful for plotting.
library(finalfit)
library(ggplot2)
theme_set(theme_bw())
explanatory = c("nodes", "extent.factor", "perfor.factor")
dependent = 'mort_5yr'
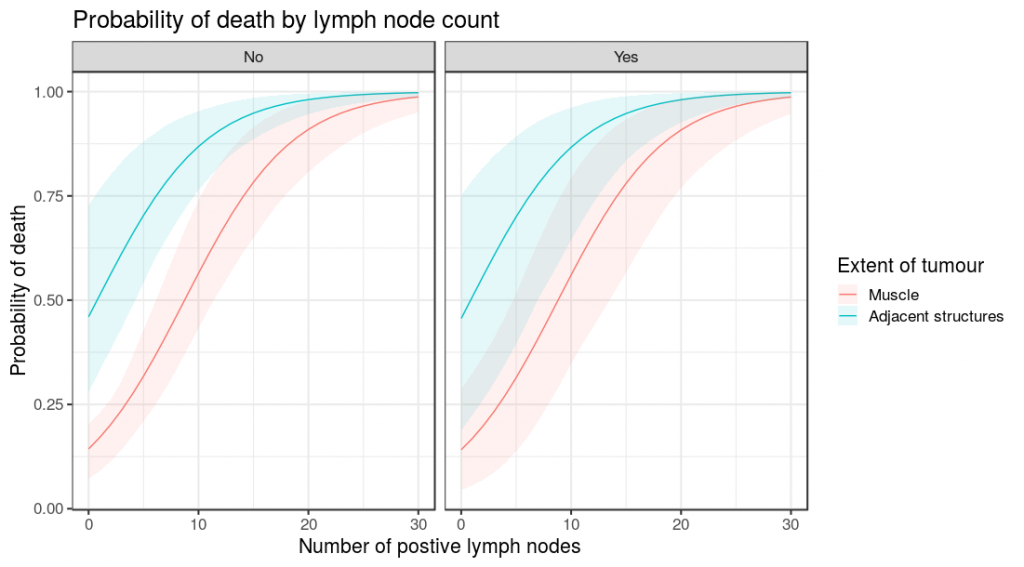
colon_s %>%
finalfit_newdata(explanatory = explanatory, rowwise = FALSE,
newdata = list(
rep(seq(0, 30), 4),
c(rep("Muscle", 62), rep("Adjacent structures", 62)),
c(rep("No", 31), rep("Yes", 31), rep("No", 31), rep("Yes", 31))
)
) -> newdata
colon_s %>%
glmmulti(dependent, explanatory) %>%
boot_predict(newdata, boot_compare = FALSE,
R=100, condense=FALSE) %>%
ggplot(aes(x = nodes, y = estimate, ymin = estimate_conf.low,
ymax = estimate_conf.high, fill=extent.factor))+
geom_line(aes(colour = extent.factor))+
geom_ribbon(alpha=0.1)+
facet_grid(.~perfor.factor)+
xlab("Number of postive lymph nodes")+
ylab("Probability of death")+
labs(fill = "Extent of tumour", colour = "Extent of tumour")+
ggtitle("Probability of death by lymph node count")So there you have it. Straightforward bootstrapped simulations of model predictions, together with comparisons and easy plotting.