Using finalfit conventions, produces a missing data matrix using
md.pattern.
missing_pattern(
.data,
dependent = NULL,
explanatory = NULL,
rotate.names = TRUE,
...
)Arguments
- .data
Data frame. Missing values must be coded
NA.- dependent
Character vector usually of length 1, name of depdendent variable.
- explanatory
Character vector of any length: name(s) of explanatory variables.
- rotate.names
Logical. Should the orientation of variable names on plot should be vertical.
- ...
pass other arguments such as
plot = TRUEtomd.pattern.
Value
A matrix with ncol(x)+1 columns, in which each row corresponds
to a missing data pattern (1=observed, 0=missing). Rows and columns are
sorted in increasing amounts of missing information. The last column and
row contain row and column counts, respectively.
Examples
library(finalfit)
library(dplyr)
explanatory = c("age.factor", "sex.factor", "obstruct.factor", "perfor.factor")
dependent = "mort_5yr"
colon_s %>%
missing_pattern(dependent, explanatory)
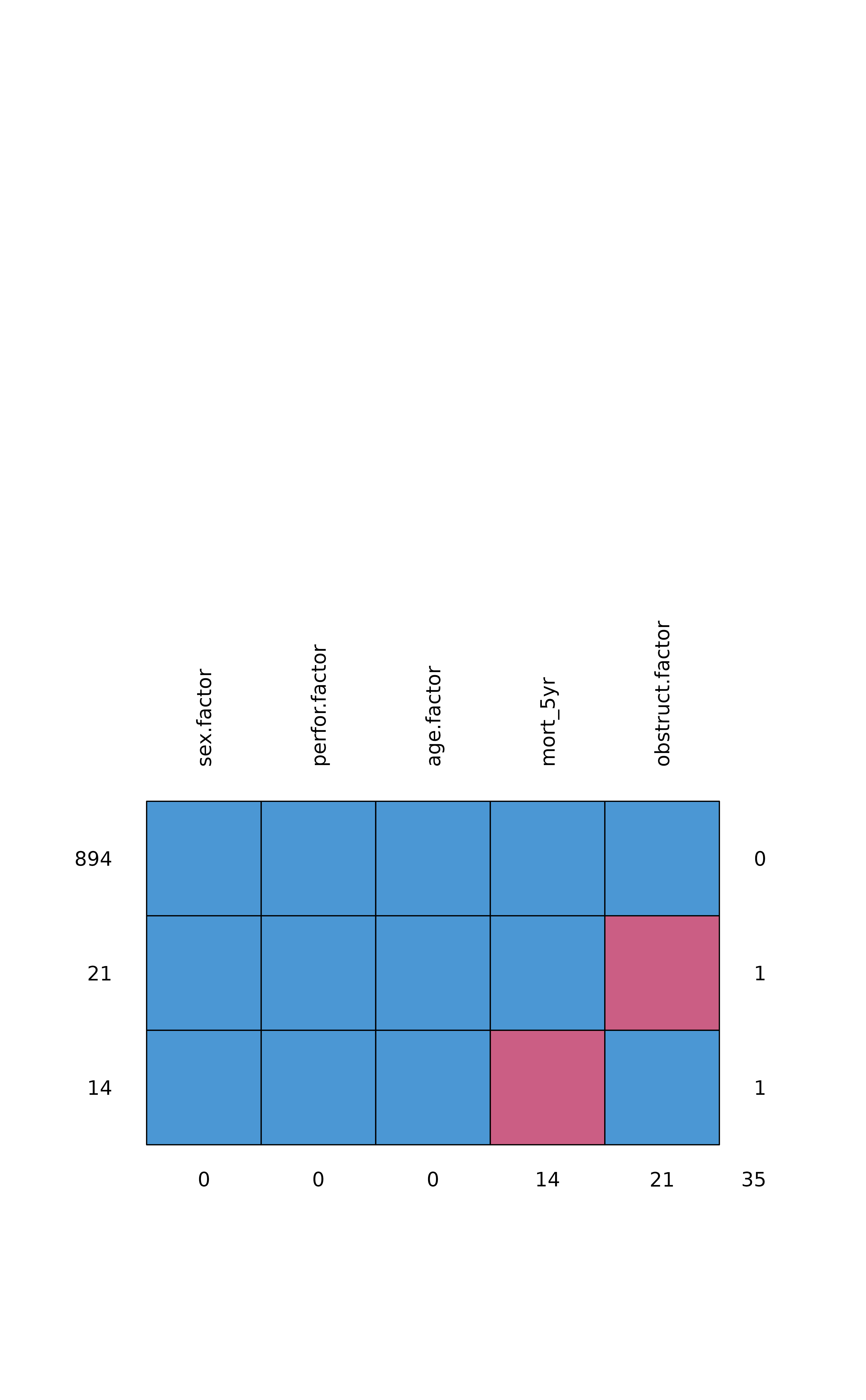 #> sex.factor perfor.factor age.factor mort_5yr obstruct.factor
#> 894 1 1 1 1 1 0
#> 21 1 1 1 1 0 1
#> 14 1 1 1 0 1 1
#> 0 0 0 14 21 35
#> sex.factor perfor.factor age.factor mort_5yr obstruct.factor
#> 894 1 1 1 1 1 0
#> 21 1 1 1 1 0 1
#> 14 1 1 1 0 1 1
#> 0 0 0 14 21 35